This is an old revision of the document!
Federated SPARQL query processing with Corese/KGRAM
Motivations
Most of scientific disciplines, and especially biomedical sciences, face nowadays “data deluge” challenges. In spite of the continuously increasing computing and storage capabilities provided by computing infrastructures, the management of massive scientific data through centralized approaches became inappropriate, for several reasons: (i) they do not guarantee the autonomy property of data providers, constrained, for either ethical or legal concerns, to keep the control over the data they host, (ii) they do not scale and adapt to the massive scientific data produced through e-Science platforms.
Federated approaches, strongly relying on distributed query processing techniques, have gained a lot of interest to adapt to the autonomy constraints of biomedical data providers. In addition, Knowledge engineering and Semantic Web technologies became popular to enhance data and knowledge sharing.
In this page, we will briefly introduce the federation extension of the Corese/KGRAM Semantic Web engine to support distributed data sources, and to address the underlying performance issues.
Demonstration
We will now demonstrate how two federate two datasets from the french national statistical institute (INSEE), namely the demographic dataset and the geographic dataset. We will basically show the content of the two datasets and distribute a query over these two endpoints. As a result, we will show the provenance of intermediate results as well as we will monitor the cost of distributed query processing in terms of the volume of issued intermediate requests and transferred intermediate results.
The last stable version of Corese-server can be downloaded here ftp://ftp-sop.inria.fr/wimmics/soft/corese-server-3.1.3.jar, and the current development version can be found, after project compilation in trunk/kgserver/target/corese-server-3.1.4-SNAPSHOT-jar-with-dependencies.jar
Corese server launching
Here is the help description of corese-server :
java -jar corese-server-3.1.4-SNAPSHOT-jar-with-dependencies.jar -h
usage: kgserver [-e] [-h] [-l <arg>] [-p <arg>] [-v] Once launched, the server can be managed through a web user interface, available at http://localhost:<PortNumber> -e,--entailments enable RDFS entailments -h,--help print this message -l,--load <arg> data file or directory to be loaded -p,--port <arg> specify the server port -v,--version print the version information and exit
We now launch two servers on ports 9091 and 9092 we will then configure them from the web user interface two load the datasets.
java -jar corese-server-3.1.4-SNAPSHOT-jar-with-dependencies.jar -p 9091
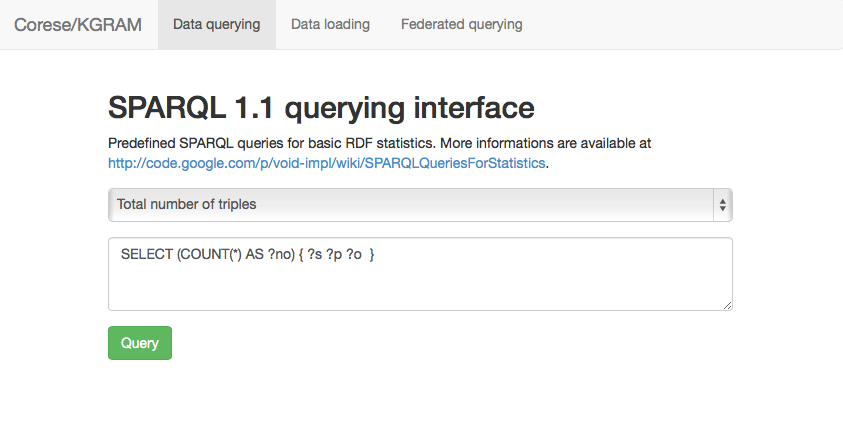
Once launched, the web querying interface can be accessed through http://localhost:9091 :
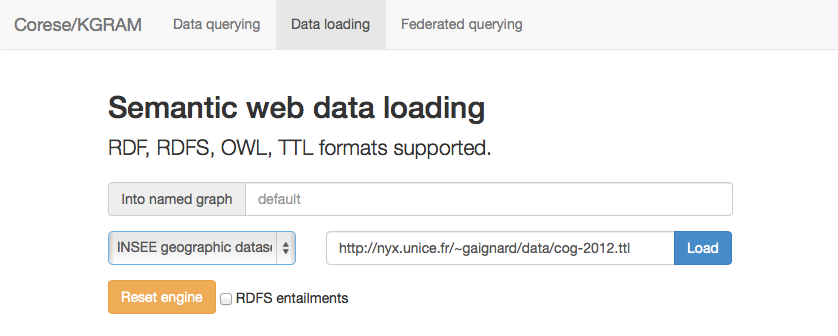
Data loading is then performed through the Data loading tab :
Finally, we can check the content served by the SPARQL endpoint through a set of statistical queries :